Welcome to the Ntranos Lab
We are developing computational methods at the intersection of information theory, genomics, and machine learning, with a particular focus on single-cell technologies and alternative splicing. Our research revolves around key algorithmic and statistical challenges that arise in computational biology — and is highly collaborative, spanning multiple biological domains in immunology, human genetics, and cancer biology.
The Ntranos lab is integrated within the broader computational research community at UCSF as part of the Dept. of Epidemiology & Biostatistics, the Diabetes Center, the Dept. of Bioengineering & Therapeutic Sciences (HIVE), and the Bakar Computational Health Sciences Institute.
Publications
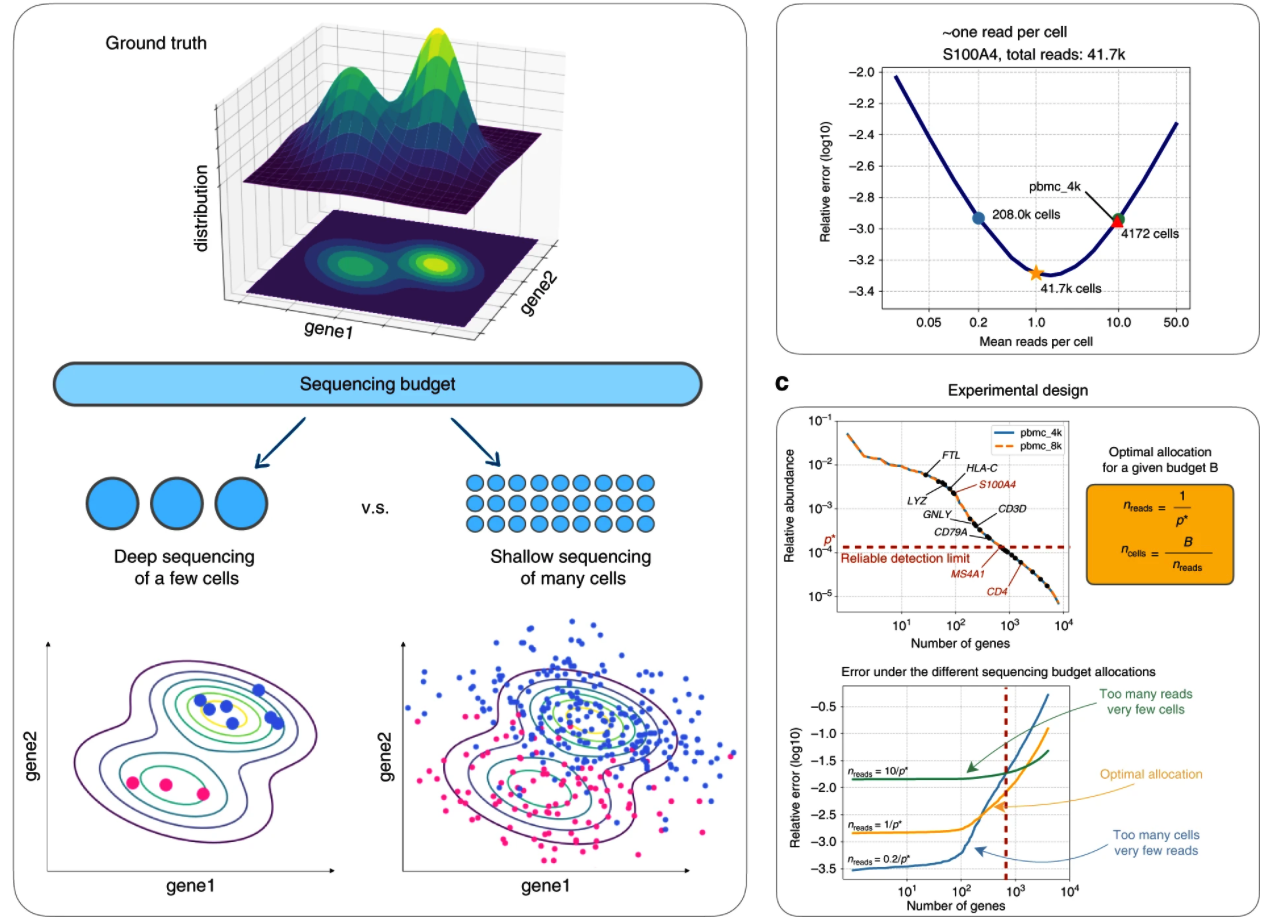
Zhang M, Ntranos V, Tse D, "Determining sequencing depth in a single-cell RNA-seq experiment,” Nature Communications, 2020
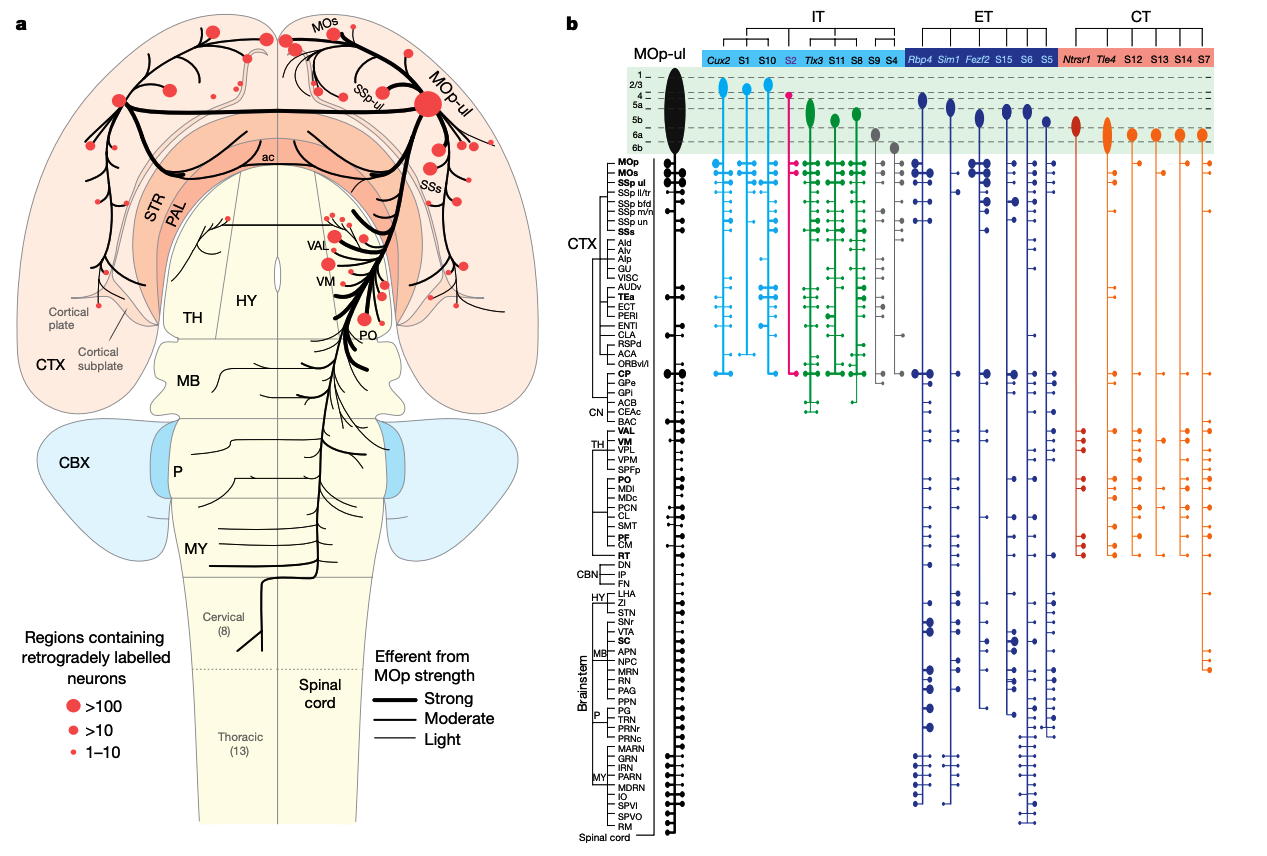
BRAIN Initiative Cell Census Network (BICCN),
"A multimodal cell census and atlas of the mammalian primary motor cortex," Nature 2021.
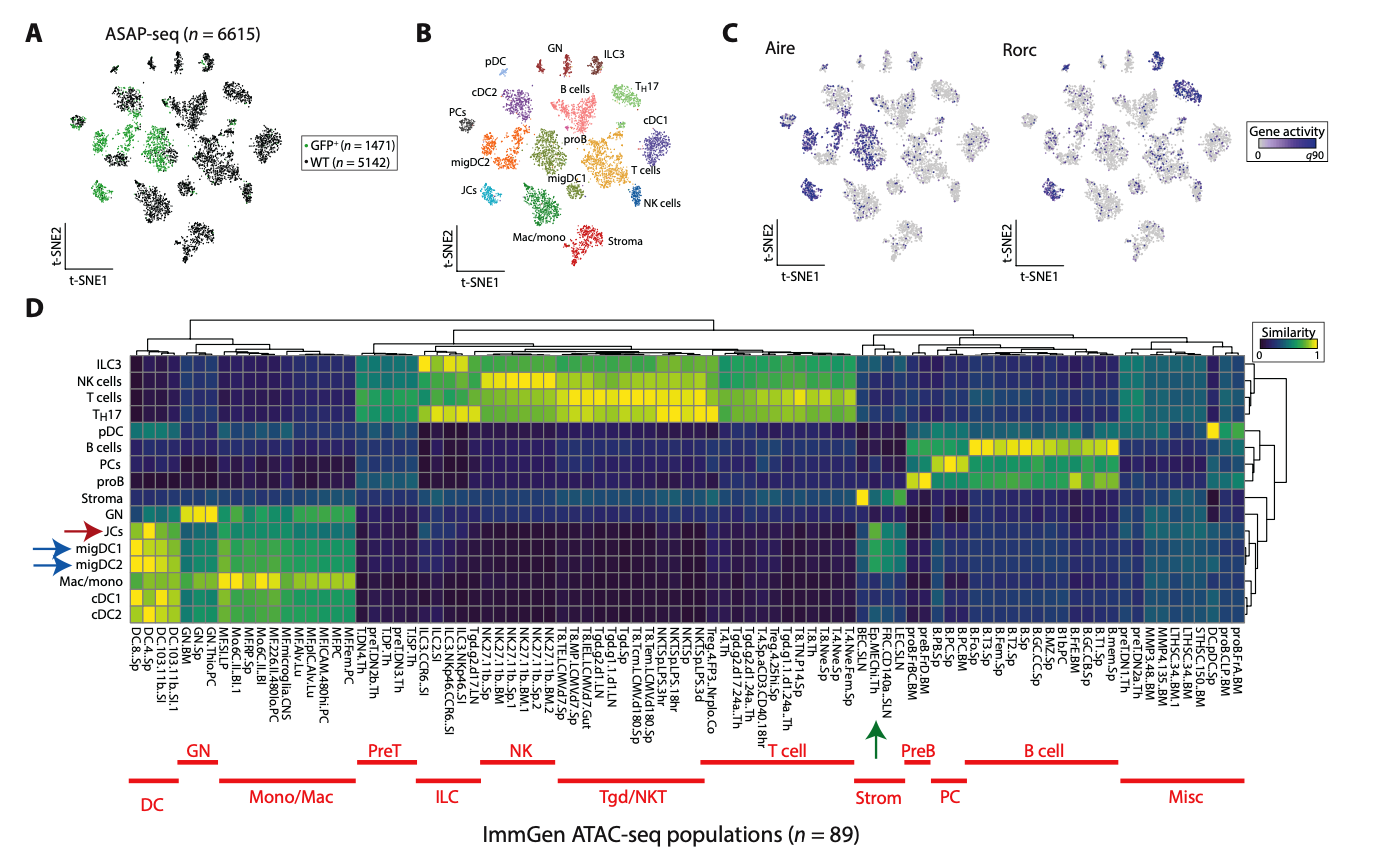
Wang J. et al., "Single-cell multiomics defines tolerogenic extrathymic Aire-expressing populations with unique homology to thymic epithelium,” Science Immunology, 2021
Selected publications
Yao et al., A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex
— Nature, 2021
Bautista et al., Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla
— Nature Communications, 2021
Gillis-Buck et al., Extrathymic Aire-expressing cells support maternal-fetal tolerance
— Science Immunology, 2021
Wang et al., Single-cell multiomics defines tolerogenic extrathymic Aire-expressing populations with unique homology to thymic epithelium
— Science Immunology, 2021
BRAIN Initiative Cell Census Network (BICCN), A multimodal cell census and atlas of the mammalian primary motor cortex
— Nature, 2021
Zhang M., Ntranos V., Tse. D, Determining sequencing depth in a single-cell RNA-seq experiment
— Nature Communications, 2020 (included in the 2020 Top 50 Life and Biological Sciences Articles collection)
Melsted P., Ntranos V., Pachter L., The barcode, UMI, set format and BUStools
— Bioinformatics, 2019
Ntranos et al., A discriminative learning approach to differential expression analysis for single-cell RNA-seq
— Nature Methods, 2019
Thompson et al.., Targeted elimination of senescent beta cells prevents type 1 diabetes
— Cell Metabolism, 2019

The view from our lab located on the 12th floor of the UCSF Parnassus Heights Medical Sciences Building
Join Us
We are hiring at all levels.
If you are interested in joining the lab please contact vasilis.ntranos at ucsf dot edu